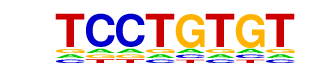
Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer
| Match Rank: | 1 |
| Score: | 0.76
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | ACACAGGA---
-AACAGGAAGT |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 2 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ACACAGGA
-CACAGN- |
|


|
|
MA0098.2_Ets1/Jaspar
| Match Rank: | 3 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACACAGGA------
NNNACAGGAAGTGGN |
|


|
|
EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 4 |
| Score: | 0.71
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | ACACAGGA---
-NACAGGAAAT |
|


|
|
ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer
| Match Rank: | 5 |
| Score: | 0.71
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ACACAGGA----
--ACAGGAAGTG |
|


|
|
MA0143.3_Sox2/Jaspar
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACACAGGA
AACAAAGG- |
|


|
|
PB0166.1_Sox12_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------ACACAGGA--
AAACAGACAAAGGAAT |
|


|
|
MA0474.1_Erg/Jaspar
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ACACAGGA-----
--ACAGGAAGTGG |
|


|
|
EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ACACAGGA---
-NACAGGAAAT |
|


|
|
MA0442.1_SOX10/Jaspar
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACACAGGA
ACAAAG-- |
|


|
|