PB0028.1_Hbp1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.82
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ATTYATTC-----
NNCATTCATTCATNNN |
|


|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTYATTC----
ACATTCATGACACG |
|


|
|
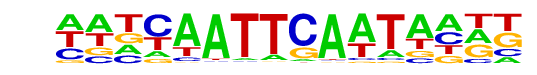
PB0068.1_Sox1_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----ATTYATTC---
AATCAATTCAATAATT |
|

|
|
MA0135.1_Lhx3/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTYATTC---
AAATTAATTAATC |
|


|
|
Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ATTYATTC
TAATTGATTA |
|


|
|
Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq(GSE14586)/Homer
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATTYATTC-
NTTTTATGAC |
|


|
|
HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTYATTC
NTATYGATCH |
|


|
|
PH0142.1_Pou1f1/Jaspar
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATTYATTC---
GANTTAATTAATTANNN |
|


|
|
HOXD13(Homeobox)/Chicken-Hoxd13-ChIP-Seq(GSE38910)/Homer
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATTYATTC--
TTTTATTRGN |
|


|
|
PH0149.1_Pou3f4/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATTYATTC---
GANTTAATTAATTAANN |
|


|
|





















