PB0148.1_Mtf1_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATTCAGAAAAAC-
AAATAAGAAAAAAC |
|


|
|
PB0182.1_Srf_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATTCAGAAAAAC-----
GTTAAAAAAAAAAATTA |
|


|
|
PB0129.1_Glis2_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATTCAGAAAAAC
AATATTAATAAAGA- |
|


|
|
PB0170.1_Sox17_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------ATTCAGAAAAAC
GACCACATTCATACAAT- |
|


|
|
PB0062.1_Sox12_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATTCAGAAAAAC--
NTTNAGAACAATTA |
|


|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTCAGAAAAAC
ACATTCATGACACG |
|


|
|
PB0192.1_Tcfap2e_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATTCAGAAAAAC-
TACTGGAAAAAAAA |
|


|
|
PH0064.1_Hoxb9/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTCAGAAAAAC--
AGAGCCATAAAATTCG |
|


|
|
PH0078.1_Hoxd13/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTCAGAAAAAC--
CTACCAATAAAATTCT |
|


|
|
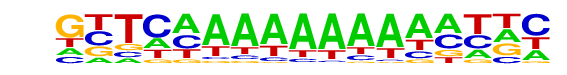
PB0116.1_Elf3_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATTCAGAAAAAC-----
GTTCAAAAAAAAAATTC |
|

|
|





















