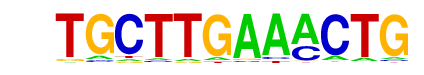
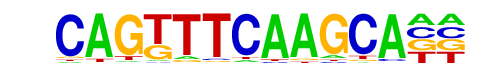
CHR/Cell-Cycle-Exp/Homer
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CAGTTTCAAGCA
CGGTTTCAAA-- |
|


|
|
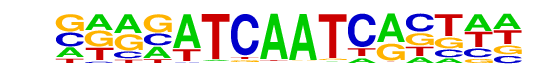
PB0188.1_Tcf7l2_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CAGTTTCAAGCA----
GAAGATCAATCACTAA |
|

|
|
PB0144.1_Lef1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CAGTTTCAAGCA----
GAAGATCAATCACTTA |
|


|
|
IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CAGTTTCAAGCA
TGGTTTCAGT-- |
|


|
|
PB0170.1_Sox17_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAGTTTCAAGCA--
GACCACATTCATACAAT |
|


|
|
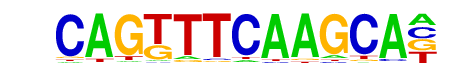
Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer
| Match Rank: | 6 |
| Score: | 0.53
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CAGTTTCAAGCA-
---TBGCACGCAA |
|

|
|
MA0069.1_Pax6/Jaspar
| Match Rank: | 7 |
| Score: | 0.52
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CAGTTTCAAGCA------
----TTCACGCATGAGTT |
|


|
|
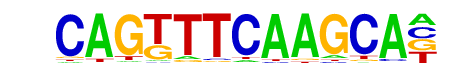
MA0144.2_STAT3/Jaspar
| Match Rank: | 8 |
| Score: | 0.52
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CAGTTTCAAGCA-
--TTTCCCAGAAN |
|

|
|
MA0070.1_PBX1/Jaspar
| Match Rank: | 9 |
| Score: | 0.52
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CAGTTTCAAGCA--
--CCATCAATCAAA |
|

|
|
PH0111.1_Nkx2-2/Jaspar
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CAGTTTCAAGCA-----
NANTTTCAAGTGGTTAN |
|


|
|