MA0078.1_Sox17/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGCAATGC-
GACAATGNN |
|


|
|
MA0019.1_Ddit3::Cebpa/Jaspar
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GGCAATGC-
AGATGCAATCCC |
|


|
|
PB0091.1_Zbtb3_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GGCAATGC------
AATCGCACTGCATTCCG |
|


|
|
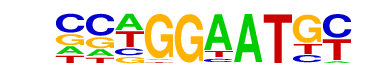
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGCAATGC
CCWGGAATGY |
|

|
|
MA0515.1_Sox6/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGCAATGC
AAAACAATGG |
|


|
|
MA0107.1_RELA/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGCAATGC--
GGAAATTCCC |
|


|
|
MA0105.3_NFKB1/Jaspar
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGCAATGC--
GGGAAATTCCC |
|


|
|
MA0597.1_THAP1/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GGCAATGC
TNNGGGCAG--- |
|


|
|
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGCAATGC
NCTGGAATGC |
|


|
|
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGCAATGC
CTTGGCAA--- |
|


|
|





















