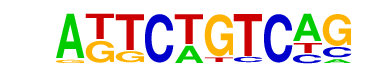
MA0498.1_Meis1/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CTGACAGAMT
NNNTGAGTGACAGCT- |
|


|
|
Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CTGACAGAMT
AGGTGHCAGACA |
|


|
|
Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTGACAGAMT
CCAGACAG--- |
|


|
|
MA0513.1_SMAD2::SMAD3::SMAD4/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTGACAGAMT-
AGGTGNCAGACAG |
|


|
|
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CTGACAGAMT
TGAGTGACAGSC- |
|


|
|
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CTGACAGAMT
GGTGYTGACAGS-- |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CTGACAGAMT
--CACAGN-- |
|


|
|
PB0028.1_Hbp1_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTGACAGAMT---
ACTATGAATGAATGAT |
|


|
|
Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 9 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTGACAGAMT
CCAGACRSVB- |
|


|
|
PH0169.1_Tgif1/Jaspar
| Match Rank: | 10 |
| Score: | 0.59
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CTGACAGAMT---
GATATTGACAGCTGCGT |
|


|
|