TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTCCATTCCA--
--GCATTCCAGN |
|


|
|
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTCCATTCCA--
--RCATTCCWGG |
|


|
|
NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTCCATTCCA
ATTTTCCATT--- |
|


|
|
MA0090.1_TEAD1/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TTCCATTCCA---
-CACATTCCTCCG |
|


|
|
PB0098.1_Zfp410_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTCCATTCCA-----
NNNTCCATCCCATAANN |
|


|
|
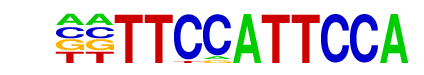
PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTCCATTCCA
GTTTCACTTCCG |
|

|
|
PB0033.1_Irf3_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTCCATTCCA
CAGTTTCGNTTCTN |
|


|
|
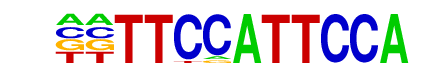
MA0152.1_NFATC2/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTCCATTCCA
TTTTCCA----- |
|

|
|
Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTCCATTCCA----
--SSAATCCACANN |
|


|
|
PB0169.1_Sox15_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TTCCATTCCA
TNGAATTTCATTNAN |
|


|
|





















