HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | reverse strand |
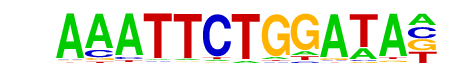
| Alignment: | ----AAATTCTGGATA
TAGAANVTTCTAGAA- |
|


|
|
PB0139.1_Irf5_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAATTCTGGATA--
NNAATTCTCGNTNAN |
|


|
|
MA0486.1_HSF1/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AAATTCTGGATA
AGAANNTTCTAGAAN |
|


|
|
PB0162.1_Sfpi1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAATTCTGGATA-
CAAATTCCGGAACC |
|


|
|
PH0148.1_Pou3f3/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAATTCTGGATA-----
AAAATATGCATAATAAA |
|


|
|
MA0092.1_Hand1::Tcfe2a/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAATTCTGGATA
--GGTCTGGCAT |
|


|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAATTCTGGATA--
ACATTCATGACACG |
|


|
|
MA0519.1_Stat5a::Stat5b/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AAATTCTGGATA--
---TTCTTGGAAAN |
|


|
|
MA0144.2_STAT3/Jaspar
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAATTCTGGATA-
--CTTCTGGGAAA |
|

|
|
PB0090.1_Zbtb12_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAATTCTGGATA--
CTAAGGTTCTAGATCAC |
|


|
|





















