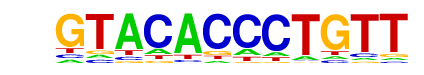
| p-value: | 1e-10 |
| log p-value: | -2.333e+01 |
| Information Content per bp: | 1.592 |
| Number of Target Sequences with motif | 11.0 |
| Percentage of Target Sequences with motif | 0.44% |
| Number of Background Sequences with motif | 1.9 |
| Percentage of Background Sequences with motif | 0.05% |
| Average Position of motif in Targets | 325.9 +/- 247.5bp |
| Average Position of motif in Background | 597.7 +/- 297.1bp |
| Strand Bias (log2 ratio + to - strand density) | 1.5 |
| Multiplicity (# of sites on avg that occur together) | 1.36 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AACAGGGTGTAC--
GVACAGNMTGTYCTB |
|


|
|
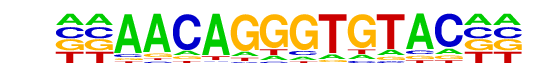
ARE(NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AACAGGGTGTAC--
AGNACAGNCTGTTCTN |
|

|
|
GRE/RAW264.7-GRE-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AACAGGGTGTAC--
GRACAGWMTGTYCTB |
|


|
|
MA0113.2_NR3C1/Jaspar
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AACAGGGTGTAC-
AGAACAGAATGTTCT |
|


|
|
MA0007.2_AR/Jaspar
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AACAGGGTGTAC
AAGAACAGAATGTTC |
|


|
|
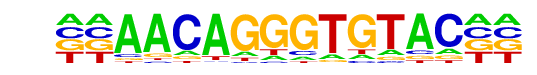
GRE(NR/IR3)/A549-GR-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AACAGGGTGTAC--
AGNACANNNTGTNCTN |
|

|
|
Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AACAGGGTGTAC
AACAGGAAGT-- |
|


|
|
SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AACAGGGTGTAC
ANCAGGATGT-- |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AACAGGGTGTAC
CACAGN------ |
|


|
|
MA0493.1_Klf1/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AACAGGGTGTAC--
---TGGGTGTGGCN |
|


|
|