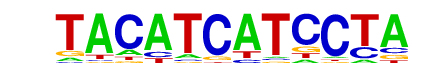
| p-value: | 1e-11 |
| log p-value: | -2.615e+01 |
| Information Content per bp: | 1.625 |
| Number of Target Sequences with motif | 15.0 |
| Percentage of Target Sequences with motif | 7.14% |
| Number of Background Sequences with motif | 1.6 |
| Percentage of Background Sequences with motif | 0.98% |
| Average Position of motif in Targets | 387.9 +/- 405.7bp |
| Average Position of motif in Background | 183.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 1.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0488.1_JUN/Jaspar
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TAGGATGATGTA--
-AAGATGATGTCAT |
|


|
|
MA0492.1_JUND_(var.2)/Jaspar
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TAGGATGATGTA---
AAAGATGATGTCATC |
|


|
|
PB0181.1_Spdef_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TAGGATGATGTA
CTACTAGGATGTNNTN |
|


|
|
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 4 |
| Score: | 0.67
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TAGGATGATGTA--
----ATGATGCAAT |
|


|
|
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | TAGGATGATGTA--
----MTGATGCAAT |
|


|
|
c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TAGGATGATGTA--
--NGATGACGTCAT |
|


|
|
MA0596.1_SREBF2/Jaspar
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TAGGATGATGTA
ATGGGGTGAT--- |
|


|
|
PH0134.1_Pbx1/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TAGGATGATGTA---
NNNNNATTGATGNGTGN |
|


|
|
ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TAGGATGATGTA
ACAGGATGTGGT- |
|


|
|
PH0037.1_Hdx/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TAGGATGATGTA-----
TNNNATGATTTCNNCNN |
|


|
|