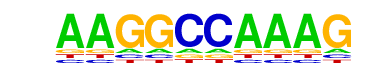
| p-value: | 1e-11 |
| log p-value: | -2.729e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 26.0 |
| Percentage of Target Sequences with motif | 7.93% |
| Number of Background Sequences with motif | 4.8 |
| Percentage of Background Sequences with motif | 1.64% |
| Average Position of motif in Targets | 437.8 +/- 467.3bp |
| Average Position of motif in Background | 746.8 +/- 455.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
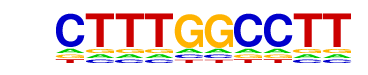
| PDF Format Logos: | forward logo
reverse opposite |
MA0484.1_HNF4G/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAGGCCAAAG----
AGAGTCCAAAGTCCA |
|


|
|
HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AAGGCCAAAG----
CANAGNNCAAAGTCCA |
|


|
|
MA0114.2_HNF4A/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAGGCCAAAG-----
NAGNNCAAAGTCCAN |
|


|
|
Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAGGCCAAAG
NTCAAGGTCA--- |
|


|
|
Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAGGCCAAAG
BTCAAGGTCA--- |
|


|
|
MA0160.1_NR4A2/Jaspar
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAGGCCAAAG
AAGGTCAC-- |
|

|
|
MA0115.1_NR1H2::RXRA/Jaspar
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAGGCCAAAG------
AAAGGTCAAAGGTCAAC |
|


|
|
RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 8 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAGGCCAAAG----
TAGGGCAAAGGTCA |
|


|
|
MA0141.2_Esrrb/Jaspar
| Match Rank: | 9 |
| Score: | 0.65
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----AAGGCCAAAG
AGCTCAAGGTCA--- |
|


|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 10 |
| Score: | 0.65
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AAGGCCAAAG
--TGCCAA-- |
|

|
|