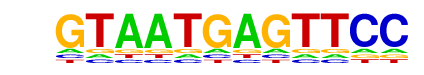
| p-value: | 1e-8 |
| log p-value: | -2.063e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 16.0 |
| Percentage of Target Sequences with motif | 7.51% |
| Number of Background Sequences with motif | 3.1 |
| Percentage of Background Sequences with motif | 1.04% |
| Average Position of motif in Targets | 374.3 +/- 340.2bp |
| Average Position of motif in Background | 392.2 +/- 368.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
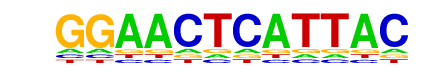
PH0151.1_Pou6f1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GTAATGAGTTCC-
GACGATAATGAGCTTGC |
|


|
|
PH0152.1_Pou6f1_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GTAATGAGTTCC-
AAACATAATGAGGTTGC |
|


|
|
PH0028.1_En1/Jaspar
| Match Rank: | 3 |
| Score: | 0.65
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GTAATGAGTTCC-
GNATTAATTAGTTNNC |
|


|
|
Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GTAATGAGTTCC
GKTAATGR----- |
|


|
|
PH0081.1_Pdx1/Jaspar
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GTAATGAGTTCC-
AAGGTAATTAGCTCAT |
|


|
|
PH0045.1_Hoxa1/Jaspar
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GTAATGAGTTCC-
ACGGTAATTAGCTCAG |
|


|
|
PH0058.1_Hoxb3/Jaspar
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GTAATGAGTTCC
TNNNACTAATTAGNTCA |
|


|
|
PH0039.1_Mnx1/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GTAATGAGTTCC-
GTACTAATTAGTGGCG |
|


|
|
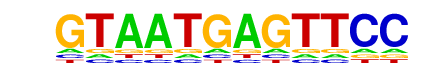
Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GTAATGAGTTCC
-TAATTAGN--- |
|

|
|
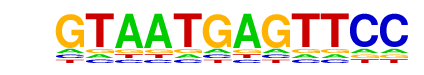
Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTAATGAGTTCC
YTAATTAVHT-- |
|

|
|