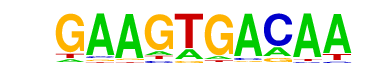
| p-value: | 1e-19 |
| log p-value: | -4.477e+01 |
| Information Content per bp: | 1.676 |
| Number of Target Sequences with motif | 34.0 |
| Percentage of Target Sequences with motif | 15.96% |
| Number of Background Sequences with motif | 6.2 |
| Percentage of Background Sequences with motif | 2.11% |
| Average Position of motif in Targets | 655.8 +/- 620.4bp |
| Average Position of motif in Background | 275.6 +/- 324.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.12 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAAGTGACAA
CGGAAGTGAAAC |
|


|
|
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 2 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GAAGTGACAA-
-AGGTGTGAAM |
|


|
|
Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GAAGTGACAA-
-AGGTGTTAAT |
|


|
|
PRDM1/BMI1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GAAGTGACAA-
GAAAGTGAAAGT |
|


|
|
PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GAAGTGACAA
AGAGGAAGTG---- |
|


|
|
MA0080.3_Spi1/Jaspar
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -8
| | Orientation: | forward strand |
| Alignment: | --------GAAGTGACAA
AAAAAGAGGAAGTGA--- |
|


|
|
MA0508.1_PRDM1/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAAGTGACAA---
AGAAAGTGAAAGTGA |
|


|
|
MF0009.1_TRP(MYB)_class/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GAAGTGACAA
-AACCGANA- |
|

|
|
ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GAAGTGACAA
ACAGGAAGTG---- |
|


|
|
MA0122.1_Nkx3-2/Jaspar
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GAAGTGACAA
TTAAGTGGA-- |
|


|
|