| p-value: | 1e-34 |
| log p-value: | -7.841e+01 |
| Information Content per bp: | 1.837 |
| Number of Target Sequences with motif | 44.0 |
| Percentage of Target Sequences with motif | 14.43% |
| Number of Background Sequences with motif | 2.2 |
| Percentage of Background Sequences with motif | 1.21% |
| Average Position of motif in Targets | 302.7 +/- 241.6bp |
| Average Position of motif in Background | 1394.4 +/- 350.9bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.18 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0130.1_ZNF354C/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CCGTGGAG
--GTGGAT |
|


|
|
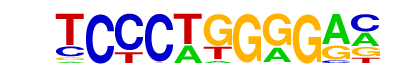
MA0154.2_EBF1/Jaspar
| Match Rank: | 2 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCGTGGAG--
TCCCTGGGGAN |
|

|
|
PB0199.1_Zfp161_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCGTGGAG-----
GCCGCGCAGTGCGT |
|


|
|
MA0259.1_HIF1A::ARNT/Jaspar
| Match Rank: | 4 |
| Score: | 0.54
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCGTGGAG
GGACGTGC-- |
|


|
|
PB0103.1_Zic3_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.53
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CCGTGGAG--
CCCCCCCGGGGGGGT |
|


|
|
PB0111.1_Bhlhb2_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.52
| | Offset: | -9
| | Orientation: | forward strand |
| Alignment: | ---------CCGTGGAG------
TGTCATTACACGTGGAAGGCGGT |
|


|
|
c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer
| Match Rank: | 7 |
| Score: | 0.51
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCGTGGAG
CACGTGGN- |
|


|
|
EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer
| Match Rank: | 8 |
| Score: | 0.51
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCGTGGAG---
TCCCNNGGGACN |
|


|
|
PB0102.1_Zic2_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.51
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CCGTGGAG--
CCCCCCCGGGGGGGT |
|


|
|
MA0006.1_Arnt::Ahr/Jaspar
| Match Rank: | 10 |
| Score: | 0.51
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCGTGGAG
TGCGTG--- |
|


|
|





















