| p-value: | 1e-9 |
| log p-value: | -2.233e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 20.0 |
| Percentage of Target Sequences with motif | 0.71% |
| Number of Background Sequences with motif | 4.5 |
| Percentage of Background Sequences with motif | 0.13% |
| Average Position of motif in Targets | 515.4 +/- 498.9bp |
| Average Position of motif in Background | 426.5 +/- 203.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0119.1_TLX1::NFIC/Jaspar
| Match Rank: | 1 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGGCACTATGCC--
TGGCACCATGCCAA |
|


|
|
NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 2 |
| Score: | 0.76
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGGCACTATGCC--
CTTGGCANNNTGCCAA |
|


|
|
Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGGCACTATGCC-
CTGGCAGNCTGCCA |
|


|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGGCACTATGCC
TTGGCA------- |
|


|
|
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGGCACTATGCC
CTTGGCAA------ |
|


|
|
ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGGCACTATGCC-
NAGGTCACNNTGACC |
|


|
|
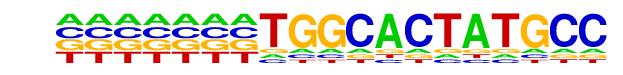
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------TGGCACTATGCC
NGTAGGTTGGCATNNN--- |
|

|
|
ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer
| Match Rank: | 8 |
| Score: | 0.53
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | TGGCACTATGCC-
-----CTAGGCCT |
|


|
|
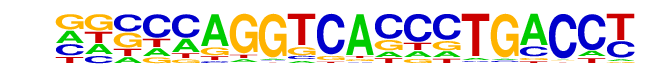
MA0112.2_ESR1/Jaspar
| Match Rank: | 9 |
| Score: | 0.52
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TGGCACTATGCC--
GGCCCAGGTCACCCTGACCT |
|

|
|
MA0258.2_ESR2/Jaspar
| Match Rank: | 10 |
| Score: | 0.51
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGGCACTATGCC--
AGGTCACCCTGACCT |
|


|
|





















