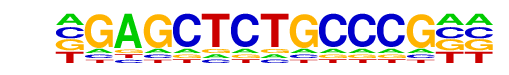
| p-value: | 1e-8 |
| log p-value: | -1.867e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 18.0 |
| Percentage of Target Sequences with motif | 0.64% |
| Number of Background Sequences with motif | 4.2 |
| Percentage of Background Sequences with motif | 0.12% |
| Average Position of motif in Targets | 468.2 +/- 408.4bp |
| Average Position of motif in Background | 823.6 +/- 242.5bp |
| Strand Bias (log2 ratio + to - strand density) | 2.0 |
| Multiplicity (# of sites on avg that occur together) | 1.11 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
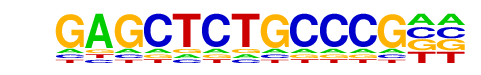
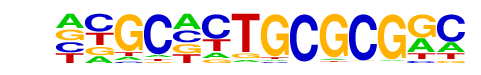
PB0199.1_Zfp161_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GAGCTCTGCCCG--
NNGCNCTGCGCGGC |
|
|
|
PB0099.1_Zfp691_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GAGCTCTGCCCG
NNNNTGAGCACTGTNNG |
|


|
|
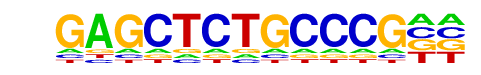
MA0597.1_THAP1/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | GAGCTCTGCCCG--
-----CTGCCCGCA |
|

|
|
RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GAGCTCTGCCCG-
TGACCTTTGCCCTA |
|


|
|
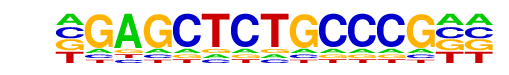
MA0504.1_NR2C2/Jaspar
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GAGCTCTGCCCG--
TGACCTCTGACCCCN |
|

|
|
PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GAGCTCTGCCCG-
TGACCTTTGCCCCA |
|


|
|
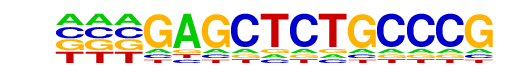
MA0065.2_PPARG::RXRA/Jaspar
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GAGCTCTGCCCG--
TGACCTTTGCCCTAN |
|

|
|
TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GAGCTCTGCCCG-
TGACCTTTGACCTC |
|


|
|
MA0512.1_Rxra/Jaspar
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GAGCTCTGCCCG
NCTGACCTTTG---- |
|

|
|
POL003.1_GC-box/Jaspar
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GAGCTCTGCCCG--
NAGCCCCGCCCCCN |
|


|
|





















