| p-value: | 1e-4 |
| log p-value: | -9.741e+00 |
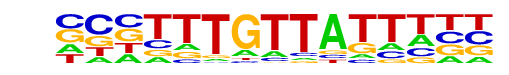
| Information Content per bp: | 1.834 |
| Number of Target Sequences with motif | 429.0 |
| Percentage of Target Sequences with motif | 15.24% |
| Number of Background Sequences with motif | 453.4 |
| Percentage of Background Sequences with motif | 12.75% |
| Average Position of motif in Targets | 428.4 +/- 392.6bp |
| Average Position of motif in Background | 448.0 +/- 491.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.17 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0182.1_Srf_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.85
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GTTTTTTTTT----
NNNNTTTTTTTTTNAAC |
|


|
|
PB0116.1_Elf3_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.77
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GTTTTTTTTT-----
GNATTTTTTTTTTGANC |
|


|
|
PB0192.1_Tcfap2e_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.76
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GTTTTTTTTT------
--TTTTTTTTCNNGTN |
|


|
|
PB0093.1_Zfp105_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GTTTTTTTTT---
NTNTTGTTGTTTGTN |
|


|
|
PB0186.1_Tcf3_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTTTTTTTTT-----
NNTTTNTTTTNGNNN |
|


|
|
MA0041.1_Foxd3/Jaspar
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GTTTTTTTTT
GAATGTTTGTTT-- |
|


|
|
MA0517.1_STAT2::STAT1/Jaspar
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GTTTTTTTTT--
TCAGTTTCATTTTCC |
|


|
|
IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GTTTTTTTTT-
RSTTTCRSTTTC |
|


|
|
MA0042.1_FOXI1/Jaspar
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GTTTTTTTTT
GGATGTTTGTTT |
|


|
|
PB0119.1_Foxa2_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GTTTTTTTTT---
NCNTTTGTTATTTNN |
|

|
|





















