| p-value: | 1e-39 |
| log p-value: | -9.170e+01 |
| Information Content per bp: | 1.809 |
| Number of Target Sequences with motif | 34.0 |
| Percentage of Target Sequences with motif | 0.92% |
| Number of Background Sequences with motif | 0.8 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 406.5 +/- 417.2bp |
| Average Position of motif in Background | 1302.6 +/- 315.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0028.1_Hbp1_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTAATGCATTCA----
NNCATTCATTCATNNN |
|


|
|
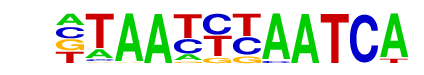
MA0468.1_DUX4/Jaspar
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTAATGCATTCA
-TAATTTAATCA |
|

|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CTAATGCATTCA-------
-----ACATTCATGACACG |
|


|
|
PB0171.1_Sox18_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTAATGCATTCA----
NNNNTGAATTCANNNC |
|


|
|
PB0068.1_Sox1_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTAATGCATTCA--
NNNTATTGAATTGNNN |
|


|
|
Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTAATGCATTCA---
CCATTGTATGCAAAT |
|


|
|
PH0014.1_Cphx/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTAATGCATTCA--
ATGATCGAATCAAA |
|


|
|
MA0132.1_Pdx1/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTAATGCATTCA
CTAATT------ |
|


|
|
Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTAATGCATTCA
-TAATTGATTA- |
|


|
|
PB0170.1_Sox17_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTAATGCATTCA------
-GACCACATTCATACAAT |
|


|
|





















