| p-value: | 1e-6 |
| log p-value: | -1.385e+01 |
| Information Content per bp: | 1.953 |
| Number of Target Sequences with motif | 26.0 |
| Percentage of Target Sequences with motif | 0.70% |
| Number of Background Sequences with motif | 9.9 |
| Percentage of Background Sequences with motif | 0.25% |
| Average Position of motif in Targets | 568.1 +/- 487.2bp |
| Average Position of motif in Background | 588.5 +/- 361.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACCGGATC--
RCCGGAARYN |
|


|
|
PB0077.1_Spdef_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----ACCGGATC---
AANNATCCGGATGTNN |
|


|
|
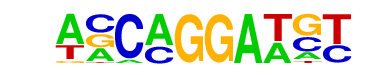
SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACCGGATC-
ANCAGGATGT |
|

|
|
MF0001.1_ETS_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACCGGATC
ACCGGAAG |
|


|
|
PB0020.1_Gabpa_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ACCGGATC-----
CAATACCGGAAGTGTAA |
|


|
|
Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACCGGATC--
DCCGGAARYN |
|


|
|
Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACCGGATC--
RCCGGAAGTD |
|


|
|
POL013.1_MED-1/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | ACCGGATC
--CGGAGC |
|


|
|
MA0028.1_ELK1/Jaspar
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ACCGGATC
GAGCCGGAAG |
|


|
|
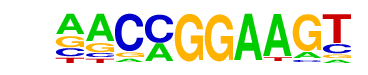
ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACCGGATC-
AACCGGAAGT |
|

|
|





















