| p-value: | 1e-16 |
| log p-value: | -3.843e+01 |
| Information Content per bp: | 1.893 |
| Number of Target Sequences with motif | 54.0 |
| Percentage of Target Sequences with motif | 1.92% |
| Number of Background Sequences with motif | 15.8 |
| Percentage of Background Sequences with motif | 0.49% |
| Average Position of motif in Targets | 403.9 +/- 386.4bp |
| Average Position of motif in Background | 527.4 +/- 474.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0119.1_Foxa2_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGTTAGWAT-
NCNTTTGTTATTTNN |
|

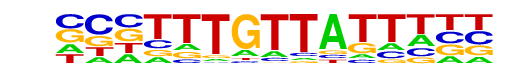
|
|
MF0011.1_HMG_class/Jaspar
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTGTTAGWAT
ATTGTT----- |
|


|
|
MA0087.1_Sox5/Jaspar
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTGTTAGWAT
ATTGTTA---- |
|


|
|
PB0172.1_Sox1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TTGTTAGWAT
CTATAATTGTTAGCG- |
|


|
|
PB0062.1_Sox12_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTGTTAGWAT-
TAATTGTTCTAAAC |
|


|
|
MA0084.1_SRY/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTGTTAGWAT
ATTGTTTAN-- |
|


|
|
STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer
| Match Rank: | 7 |
| Score: | 0.65
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTGTTAGWAT
NTTTCTNAGAAA |
|


|
|
Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTGTTAGWAT
CCATTGTTNY--- |
|


|
|
PB0123.1_Foxl1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGTTAGWAT--
NNTTTTGTTTTGATNT |
|


|
|
PB0194.1_Zbtb12_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTGTTAGWAT---
TATCATTAGAACGCT |
|


|
|





















