| p-value: | 1e-7 |
| log p-value: | -1.776e+01 |
| Information Content per bp: | 1.668 |
| Number of Target Sequences with motif | 421.0 |
| Percentage of Target Sequences with motif | 14.96% |
| Number of Background Sequences with motif | 370.2 |
| Percentage of Background Sequences with motif | 11.51% |
| Average Position of motif in Targets | 435.4 +/- 383.3bp |
| Average Position of motif in Background | 471.9 +/- 437.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.30 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0092.1_Hand1::Tcfe2a/Jaspar
| Match Rank: | 1 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTGGGAYTAC
GGTCTGGCAT--- |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTGGGAYTAC
GCTGTG----- |
|


|
|
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTGGGAYTAC
NCTGGAATGC- |
|


|
|
Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTGGGAYTAC
CSTGGGAAAD- |
|


|
|
PB0195.1_Zbtb3_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CTGGGAYTAC
CAATCACTGGCAGAAT |
|


|
|
PH0169.1_Tgif1/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CTGGGAYTAC-
NNNCAGCTGTCAATATN |
|


|
|
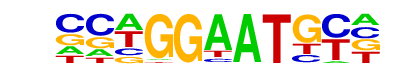
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTGGGAYTAC
CCWGGAATGY- |
|

|
|
MA0156.1_FEV/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTGGGAYTAC
CAGGAAAT-- |
|


|
|
ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CTGGGAYTAC
RGSMTBCTGGGAAAT- |
|


|
|
EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CTGGGAYTAC
NACAGGAAAT-- |
|


|
|





















