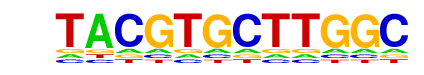
| p-value: | 1e-36 |
| log p-value: | -8.321e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 132.0 |
| Percentage of Target Sequences with motif | 0.11% |
| Number of Background Sequences with motif | 5.7 |
| Percentage of Background Sequences with motif | 0.03% |
| Average Position of motif in Targets | 531.6 +/- 469.8bp |
| Average Position of motif in Background | 672.8 +/- 278.9bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
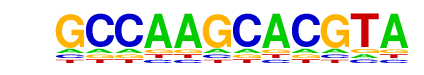
HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
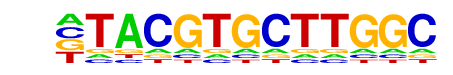
| Alignment: | TACGTGCTTGGC
TACGTGCV---- |
|

|
|
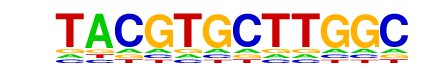
HIF2a(HLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TACGTGCTTGGC
GGGTACGTGC----- |
|

|
|
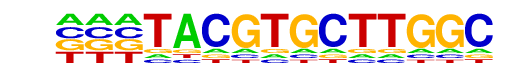
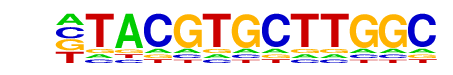
MA0259.1_HIF1A::ARNT/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TACGTGCTTGGC
GGACGTGC----- |
|

|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | TACGTGCTTGGC-
-------TTGGCA |
|


|
|
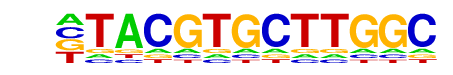
Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TACGTGCTTGGC
TTAAGTGCTT--- |
|

|
|
POL004.1_CCAAT-box/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TACGTGCTTGGC----
----TGATTGGCTANN |
|


|
|
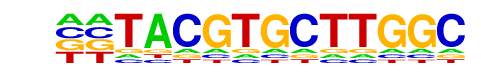
MA0059.1_MYC::MAX/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TACGTGCTTGGC
ACCACGTGCTC--- |
|

|
|
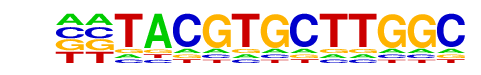
MA0464.1_Bhlhe40/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TACGTGCTTGGC
CTCACGTGCAC--- |
|

|
|
MA0004.1_Arnt/Jaspar
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TACGTGCTTGGC
CACGTG------ |
|


|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TACGTGCTTGGC
NNACTTGCCTT-- |
|

|
|