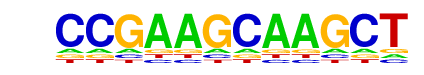
| p-value: | 1e-57 |
| log p-value: | -1.317e+02 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 165.0 |
| Percentage of Target Sequences with motif | 0.14% |
| Number of Background Sequences with motif | 5.6 |
| Percentage of Background Sequences with motif | 0.03% |
| Average Position of motif in Targets | 563.5 +/- 501.6bp |
| Average Position of motif in Background | 866.8 +/- 573.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
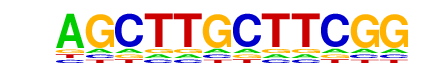
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0164.1_Nr2e3/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | CCGAAGCAAGCT-
------CAAGCTT |
|


|
|
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCGAAGCAAGCT
-NGAAGC----- |
|

|
|
PB0055.1_Rfx4_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.53
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCGAAGCAAGCT-
TACCATAGCAACGGT |
|


|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 4 |
| Score: | 0.53
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCGAAGCAAGCT-
--AAGGCAAGTGT |
|


|
|
POL013.1_MED-1/Jaspar
| Match Rank: | 5 |
| Score: | 0.52
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCGAAGCAAGCT
-CGGAGC----- |
|

|
|
PB0054.1_Rfx3_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.52
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CCGAAGCAAGCT-----
TGTGACCCTTAGCAACCGATTAA |
|


|
|
PH0026.1_Duxbl/Jaspar
| Match Rank: | 7 |
| Score: | 0.51
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCGAAGCAAGCT--
CGACCCAATCAACGGTG |
|


|
|
PB0035.1_Irf5_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.51
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CCGAAGCAAGCT
ATAAACCGAAACCAA-- |
|


|
|
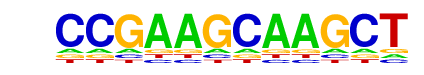
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 9 |
| Score: | 0.50
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCGAAGCAAGCT
MTGATGCAAT-- |
|

|
|
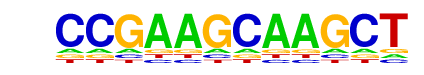
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 10 |
| Score: | 0.50
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCGAAGCAAGCT
ATGATGCAAT-- |
|

|
|