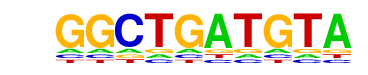
| p-value: | 1e-34 |
| log p-value: | -7.889e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 142.0 |
| Percentage of Target Sequences with motif | 0.11% |
| Number of Background Sequences with motif | 5.6 |
| Percentage of Background Sequences with motif | 0.04% |
| Average Position of motif in Targets | 459.7 +/- 390.6bp |
| Average Position of motif in Background | 661.1 +/- 734.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TACATCAGCC
ATTGCATCAT-- |
|


|
|
MA0117.1_Mafb/Jaspar
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TACATCAGCC
-NCGTCAGC- |
|


|
|
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TACATCAGCC
ATTGCATCAK-- |
|


|
|
MA0025.1_NFIL3/Jaspar
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TACATCAGCC
ANGTTACATAA--- |
|


|
|
MA0488.1_JUN/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TACATCAGCC-
ATGACATCATCNN |
|


|
|
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 6 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TACATCAGCC
-ACGTCA--- |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | TACATCAGCC
-----CAGCC |
|


|
|
Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TACATCAGCC
TGACGTCATC- |
|


|
|
PB0051.1_Osr2_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TACATCAGCC---
ATGTACAGTAGCAAAG |
|


|
|
PH0134.1_Pbx1/Jaspar
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TACATCAGCC----
TCACCCATCAATAAACA |
|


|
|