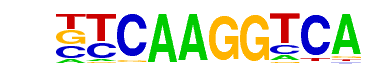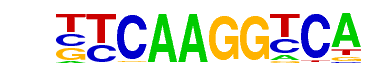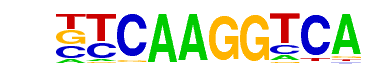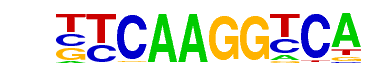| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-53 | -1.240e+02 | 0.0000 | 108.0 | 10.99% | 7.2 | 1.64% | motif file (matrix) |
pdf |
| 2 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-44 | -1.017e+02 | 0.0000 | 114.0 | 11.60% | 10.4 | 2.35% | motif file (matrix) |
pdf |
| 3 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-32 | -7.529e+01 | 0.0000 | 93.0 | 9.46% | 9.5 | 2.15% | motif file (matrix) |
pdf |
| 4 | 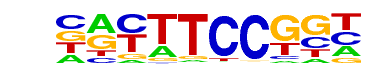 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-29 | -6.744e+01 | 0.0000 | 173.0 | 17.60% | 30.4 | 6.90% | motif file (matrix) |
pdf |
| 5 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-25 | -5.769e+01 | 0.0000 | 41.0 | 4.17% | 2.6 | 0.59% | motif file (matrix) |
pdf |
| 6 |  | ETS(ETS)/Promoter/Homer | 1e-21 | -4.874e+01 | 0.0000 | 66.0 | 6.71% | 7.2 | 1.63% | motif file (matrix) |
pdf |
| 7 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-20 | -4.740e+01 | 0.0000 | 179.0 | 18.21% | 38.2 | 8.68% | motif file (matrix) |
pdf |
| 8 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-20 | -4.705e+01 | 0.0000 | 146.0 | 14.85% | 28.3 | 6.43% | motif file (matrix) |
pdf |
| 9 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-18 | -4.329e+01 | 0.0000 | 125.0 | 12.72% | 23.7 | 5.38% | motif file (matrix) |
pdf |
| 10 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-17 | -4.117e+01 | 0.0000 | 133.0 | 13.53% | 26.7 | 6.06% | motif file (matrix) |
pdf |
| 11 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-15 | -3.652e+01 | 0.0000 | 97.0 | 9.87% | 17.6 | 3.98% | motif file (matrix) |
pdf |
| 12 |  | YY1(Zf)/Promoter/Homer | 1e-15 | -3.547e+01 | 0.0000 | 23.0 | 2.34% | 0.3 | 0.08% | motif file (matrix) |
pdf |
| 13 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-15 | -3.492e+01 | 0.0000 | 142.0 | 14.45% | 31.7 | 7.19% | motif file (matrix) |
pdf |
| 14 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-14 | -3.335e+01 | 0.0000 | 236.0 | 24.01% | 64.7 | 14.69% | motif file (matrix) |
pdf |
| 15 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-14 | -3.328e+01 | 0.0000 | 313.0 | 31.84% | 93.3 | 21.18% | motif file (matrix) |
pdf |
| 16 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-14 | -3.238e+01 | 0.0000 | 145.0 | 14.75% | 33.9 | 7.70% | motif file (matrix) |
pdf |
| 17 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-13 | -3.172e+01 | 0.0000 | 177.0 | 18.01% | 44.5 | 10.10% | motif file (matrix) |
pdf |
| 18 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-12 | -2.910e+01 | 0.0000 | 176.0 | 17.90% | 45.4 | 10.31% | motif file (matrix) |
pdf |
| 19 |  | E-box(HLH)/Promoter/Homer | 1e-12 | -2.853e+01 | 0.0000 | 20.0 | 2.03% | 0.5 | 0.11% | motif file (matrix) |
pdf |
| 20 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-12 | -2.853e+01 | 0.0000 | 20.0 | 2.03% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 21 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-11 | -2.573e+01 | 0.0000 | 57.0 | 5.80% | 9.7 | 2.20% | motif file (matrix) |
pdf |
| 22 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-10 | -2.395e+01 | 0.0000 | 267.0 | 27.16% | 82.3 | 18.69% | motif file (matrix) |
pdf |
| 23 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.252e+01 | 0.0000 | 54.0 | 5.49% | 9.3 | 2.11% | motif file (matrix) |
pdf |
| 24 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-9 | -2.221e+01 | 0.0000 | 42.0 | 4.27% | 6.6 | 1.49% | motif file (matrix) |
pdf |
| 25 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.218e+01 | 0.0000 | 68.0 | 6.92% | 13.2 | 3.00% | motif file (matrix) |
pdf |
| 26 |  | TATA-Box(TBP)/Promoter/Homer | 1e-9 | -2.159e+01 | 0.0000 | 202.0 | 20.55% | 59.6 | 13.53% | motif file (matrix) |
pdf |
| 27 |  | Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.123e+01 | 0.0000 | 77.0 | 7.83% | 16.5 | 3.75% | motif file (matrix) |
pdf |
| 28 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-8 | -1.970e+01 | 0.0000 | 193.0 | 19.63% | 57.8 | 13.11% | motif file (matrix) |
pdf |
| 29 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.960e+01 | 0.0000 | 100.0 | 10.17% | 24.1 | 5.46% | motif file (matrix) |
pdf |
| 30 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.924e+01 | 0.0000 | 68.0 | 6.92% | 14.7 | 3.34% | motif file (matrix) |
pdf |
| 31 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-8 | -1.892e+01 | 0.0000 | 102.0 | 10.38% | 25.6 | 5.82% | motif file (matrix) |
pdf |
| 32 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-8 | -1.845e+01 | 0.0000 | 116.0 | 11.80% | 30.7 | 6.98% | motif file (matrix) |
pdf |
| 33 | 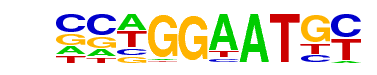 | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-7 | -1.835e+01 | 0.0000 | 110.0 | 11.19% | 28.2 | 6.41% | motif file (matrix) |
pdf |
| 34 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-7 | -1.833e+01 | 0.0000 | 295.0 | 30.01% | 98.1 | 22.27% | motif file (matrix) |
pdf |
| 35 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-7 | -1.831e+01 | 0.0000 | 206.0 | 20.96% | 63.0 | 14.30% | motif file (matrix) |
pdf |
| 36 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.758e+01 | 0.0000 | 34.0 | 3.46% | 5.9 | 1.35% | motif file (matrix) |
pdf |
| 37 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-7 | -1.714e+01 | 0.0000 | 45.0 | 4.58% | 8.6 | 1.96% | motif file (matrix) |
pdf |
| 38 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-7 | -1.702e+01 | 0.0000 | 25.0 | 2.54% | 3.4 | 0.76% | motif file (matrix) |
pdf |
| 39 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-7 | -1.644e+01 | 0.0000 | 33.0 | 3.36% | 5.6 | 1.27% | motif file (matrix) |
pdf |
| 40 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-6 | -1.610e+01 | 0.0000 | 140.0 | 14.24% | 40.2 | 9.11% | motif file (matrix) |
pdf |
| 41 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-6 | -1.608e+01 | 0.0000 | 14.0 | 1.42% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 42 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-6 | -1.606e+01 | 0.0000 | 242.0 | 24.62% | 79.9 | 18.13% | motif file (matrix) |
pdf |
| 43 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-6 | -1.559e+01 | 0.0000 | 82.0 | 8.34% | 20.5 | 4.65% | motif file (matrix) |
pdf |
| 44 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-6 | -1.551e+01 | 0.0000 | 179.0 | 18.21% | 55.2 | 12.52% | motif file (matrix) |
pdf |
| 45 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-6 | -1.505e+01 | 0.0000 | 69.0 | 7.02% | 16.1 | 3.65% | motif file (matrix) |
pdf |
| 46 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529/Homer | 1e-6 | -1.494e+01 | 0.0000 | 84.0 | 8.55% | 21.8 | 4.94% | motif file (matrix) |
pdf |
| 47 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-6 | -1.482e+01 | 0.0000 | 56.0 | 5.70% | 12.6 | 2.85% | motif file (matrix) |
pdf |
| 48 | 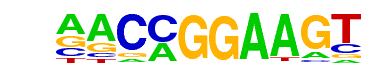 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-6 | -1.478e+01 | 0.0000 | 180.0 | 18.31% | 56.8 | 12.88% | motif file (matrix) |
pdf |
| 49 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-6 | -1.453e+01 | 0.0000 | 126.0 | 12.82% | 36.9 | 8.37% | motif file (matrix) |
pdf |
| 50 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-6 | -1.422e+01 | 0.0000 | 13.0 | 1.32% | 2.0 | 0.45% | motif file (matrix) |
pdf |
| 51 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-6 | -1.422e+01 | 0.0000 | 13.0 | 1.32% | 2.0 | 0.45% | motif file (matrix) |
pdf |
| 52 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-5 | -1.307e+01 | 0.0000 | 60.0 | 6.10% | 14.3 | 3.25% | motif file (matrix) |
pdf |
| 53 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.306e+01 | 0.0000 | 101.0 | 10.27% | 28.8 | 6.54% | motif file (matrix) |
pdf |
| 54 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-5 | -1.285e+01 | 0.0000 | 47.0 | 4.78% | 10.1 | 2.29% | motif file (matrix) |
pdf |
| 55 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-5 | -1.264e+01 | 0.0000 | 103.0 | 10.48% | 29.8 | 6.76% | motif file (matrix) |
pdf |
| 56 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-5 | -1.243e+01 | 0.0000 | 12.0 | 1.22% | 1.3 | 0.29% | motif file (matrix) |
pdf |
| 57 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-5 | -1.160e+01 | 0.0000 | 136.0 | 13.84% | 42.2 | 9.58% | motif file (matrix) |
pdf |
| 58 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-4 | -1.117e+01 | 0.0001 | 72.0 | 7.32% | 19.7 | 4.47% | motif file (matrix) |
pdf |
| 59 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.073e+01 | 0.0001 | 11.0 | 1.12% | 2.0 | 0.45% | motif file (matrix) |
pdf |
| 60 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-4 | -1.073e+01 | 0.0001 | 11.0 | 1.12% | 0.9 | 0.20% | motif file (matrix) |
pdf |
| 61 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.073e+01 | 0.0001 | 11.0 | 1.12% | 0.8 | 0.18% | motif file (matrix) |
pdf |
| 62 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-4 | -1.040e+01 | 0.0001 | 59.0 | 6.00% | 15.5 | 3.51% | motif file (matrix) |
pdf |
| 63 |  | c-Myc/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.038e+01 | 0.0001 | 56.0 | 5.70% | 14.1 | 3.21% | motif file (matrix) |
pdf |
| 64 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.028e+01 | 0.0001 | 101.0 | 10.27% | 30.2 | 6.84% | motif file (matrix) |
pdf |
| 65 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-4 | -9.886e+00 | 0.0002 | 198.0 | 20.14% | 68.4 | 15.52% | motif file (matrix) |
pdf |
| 66 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.832e+00 | 0.0002 | 137.0 | 13.94% | 44.7 | 10.15% | motif file (matrix) |
pdf |
| 67 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-4 | -9.750e+00 | 0.0002 | 559.0 | 56.87% | 223.6 | 50.76% | motif file (matrix) |
pdf |
| 68 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-4 | -9.370e+00 | 0.0003 | 284.0 | 28.89% | 104.1 | 23.64% | motif file (matrix) |
pdf |
| 69 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-4 | -9.276e+00 | 0.0003 | 88.0 | 8.95% | 26.2 | 5.95% | motif file (matrix) |
pdf |
| 70 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -9.106e+00 | 0.0004 | 10.0 | 1.02% | 1.3 | 0.29% | motif file (matrix) |
pdf |
| 71 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-3 | -8.613e+00 | 0.0006 | 56.0 | 5.70% | 15.2 | 3.45% | motif file (matrix) |
pdf |
| 72 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -8.513e+00 | 0.0007 | 105.0 | 10.68% | 33.6 | 7.63% | motif file (matrix) |
pdf |
| 73 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-3 | -8.494e+00 | 0.0007 | 18.0 | 1.83% | 3.4 | 0.77% | motif file (matrix) |
pdf |
| 74 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-3 | -8.490e+00 | 0.0007 | 414.0 | 42.12% | 161.3 | 36.63% | motif file (matrix) |
pdf |
| 75 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-3 | -8.395e+00 | 0.0007 | 25.0 | 2.54% | 5.9 | 1.35% | motif file (matrix) |
pdf |
| 76 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-3 | -7.963e+00 | 0.0011 | 31.0 | 3.15% | 7.8 | 1.78% | motif file (matrix) |
pdf |
| 77 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.949e+00 | 0.0011 | 77.0 | 7.83% | 23.2 | 5.27% | motif file (matrix) |
pdf |
| 78 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-3 | -7.911e+00 | 0.0012 | 85.0 | 8.65% | 26.5 | 6.02% | motif file (matrix) |
pdf |
| 79 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-3 | -7.874e+00 | 0.0012 | 152.0 | 15.46% | 52.5 | 11.92% | motif file (matrix) |
pdf |
| 80 |  | Sp1(Zf)/Promoter/Homer | 1e-3 | -7.825e+00 | 0.0012 | 43.0 | 4.37% | 11.3 | 2.58% | motif file (matrix) |
pdf |
| 81 |  | E2F(E2F)/Cell-Cycle-Exp/Homer | 1e-3 | -7.576e+00 | 0.0016 | 9.0 | 0.92% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 82 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-3 | -7.567e+00 | 0.0016 | 113.0 | 11.50% | 37.7 | 8.56% | motif file (matrix) |
pdf |
| 83 | 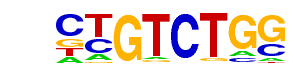 | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.550e+00 | 0.0016 | 146.0 | 14.85% | 50.6 | 11.50% | motif file (matrix) |
pdf |
| 84 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.468e+00 | 0.0017 | 17.0 | 1.73% | 3.4 | 0.78% | motif file (matrix) |
pdf |
| 85 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-3 | -7.443e+00 | 0.0017 | 265.0 | 26.96% | 99.1 | 22.50% | motif file (matrix) |
pdf |
| 86 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-3 | -7.316e+00 | 0.0019 | 120.0 | 12.21% | 40.3 | 9.14% | motif file (matrix) |
pdf |
| 87 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -7.276e+00 | 0.0020 | 197.0 | 20.04% | 71.2 | 16.17% | motif file (matrix) |
pdf |
| 88 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-3 | -7.181e+00 | 0.0021 | 33.0 | 3.36% | 8.6 | 1.96% | motif file (matrix) |
pdf |
| 89 |  | Hoxc9(Homeobox)/Ainv15-Hoxc9-ChIP-Seq(GSE21812)/Homer | 1e-2 | -6.579e+00 | 0.0038 | 100.0 | 10.17% | 33.4 | 7.58% | motif file (matrix) |
pdf |
| 90 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.523e+00 | 0.0040 | 32.0 | 3.26% | 8.8 | 1.99% | motif file (matrix) |
pdf |
| 91 | 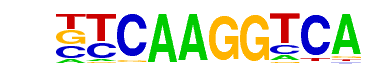 | Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer | 1e-2 | -6.360e+00 | 0.0047 | 76.0 | 7.73% | 24.3 | 5.51% | motif file (matrix) |
pdf |
| 92 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.335e+00 | 0.0047 | 179.0 | 18.21% | 65.9 | 14.96% | motif file (matrix) |
pdf |
| 93 | 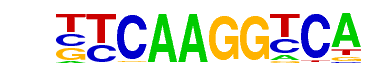 | Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer | 1e-2 | -6.247e+00 | 0.0051 | 57.0 | 5.80% | 17.2 | 3.91% | motif file (matrix) |
pdf |
| 94 |  | Pax7-longest(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-2 | -6.147e+00 | 0.0055 | 8.0 | 0.81% | 1.3 | 0.29% | motif file (matrix) |
pdf |
| 95 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-2 | -6.147e+00 | 0.0055 | 8.0 | 0.81% | 1.8 | 0.42% | motif file (matrix) |
pdf |
| 96 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-2 | -6.147e+00 | 0.0055 | 8.0 | 0.81% | 1.3 | 0.30% | motif file (matrix) |
pdf |
| 97 |  | ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER | 1e-2 | -6.114e+00 | 0.0056 | 12.0 | 1.22% | 2.8 | 0.65% | motif file (matrix) |
pdf |
| 98 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-2 | -6.069e+00 | 0.0058 | 70.0 | 7.12% | 22.7 | 5.15% | motif file (matrix) |
pdf |
| 99 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-2 | -6.059e+00 | 0.0058 | 156.0 | 15.87% | 56.9 | 12.91% | motif file (matrix) |
pdf |
| 100 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-2 | -5.897e+00 | 0.0068 | 25.0 | 2.54% | 6.5 | 1.48% | motif file (matrix) |
pdf |
| 101 |  | NRF1/Promoter/Homer | 1e-2 | -5.877e+00 | 0.0068 | 28.0 | 2.85% | 7.3 | 1.66% | motif file (matrix) |
pdf |
| 102 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-2 | -5.765e+00 | 0.0076 | 172.0 | 17.50% | 63.6 | 14.44% | motif file (matrix) |
pdf |
| 103 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-2 | -5.455e+00 | 0.0102 | 84.0 | 8.55% | 28.4 | 6.45% | motif file (matrix) |
pdf |
| 104 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.387e+00 | 0.0108 | 94.0 | 9.56% | 32.3 | 7.34% | motif file (matrix) |
pdf |
| 105 | 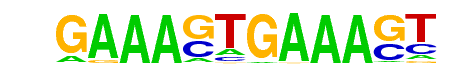 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-2 | -5.347e+00 | 0.0112 | 18.0 | 1.83% | 4.9 | 1.11% | motif file (matrix) |
pdf |
| 106 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.254e+00 | 0.0121 | 170.0 | 17.29% | 63.5 | 14.41% | motif file (matrix) |
pdf |
| 107 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.252e+00 | 0.0121 | 225.0 | 22.89% | 86.6 | 19.66% | motif file (matrix) |
pdf |
| 108 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.155e+00 | 0.0131 | 507.0 | 51.58% | 209.7 | 47.60% | motif file (matrix) |
pdf |
| 109 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -5.122e+00 | 0.0135 | 518.0 | 52.70% | 214.8 | 48.75% | motif file (matrix) |
pdf |
| 110 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-2 | -5.110e+00 | 0.0135 | 83.0 | 8.44% | 28.7 | 6.52% | motif file (matrix) |
pdf |
| 111 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-2 | -4.962e+00 | 0.0155 | 266.0 | 27.06% | 104.3 | 23.67% | motif file (matrix) |
pdf |
| 112 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.925e+00 | 0.0160 | 35.0 | 3.56% | 10.9 | 2.46% | motif file (matrix) |
pdf |
| 113 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -4.839e+00 | 0.0172 | 249.0 | 25.33% | 97.2 | 22.06% | motif file (matrix) |
pdf |
| 114 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.829e+00 | 0.0173 | 7.0 | 0.71% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 115 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-2 | -4.812e+00 | 0.0174 | 321.0 | 32.66% | 128.8 | 29.24% | motif file (matrix) |
pdf |
| 116 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-2 | -4.810e+00 | 0.0174 | 43.0 | 4.37% | 13.8 | 3.13% | motif file (matrix) |
pdf |
| 117 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-2 | -4.769e+00 | 0.0178 | 168.0 | 17.09% | 63.7 | 14.47% | motif file (matrix) |
pdf |
| 118 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-2 | -4.749e+00 | 0.0181 | 102.0 | 10.38% | 36.2 | 8.22% | motif file (matrix) |
pdf |
| 119 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-2 | -4.741e+00 | 0.0181 | 151.0 | 15.36% | 56.6 | 12.84% | motif file (matrix) |
pdf |
| 120 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-2 | -4.682e+00 | 0.0190 | 56.0 | 5.70% | 18.5 | 4.19% | motif file (matrix) |
pdf |