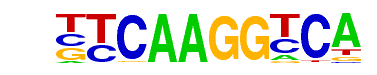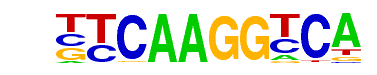| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Sp1(Zf)/Promoter/Homer | 1e-98 | -2.258e+02 | 0.0000 | 194.0 | 8.65% | 9.5 | 1.27% | motif file (matrix) |
pdf |
| 2 |  | YY1(Zf)/Promoter/Homer | 1e-78 | -1.803e+02 | 0.0000 | 77.0 | 3.43% | 0.8 | 0.11% | motif file (matrix) |
pdf |
| 3 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-74 | -1.708e+02 | 0.0000 | 281.0 | 12.53% | 26.2 | 3.50% | motif file (matrix) |
pdf |
| 4 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-67 | -1.545e+02 | 0.0000 | 69.0 | 3.08% | 0.3 | 0.04% | motif file (matrix) |
pdf |
| 5 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-56 | -1.297e+02 | 0.0000 | 327.0 | 14.59% | 41.1 | 5.49% | motif file (matrix) |
pdf |
| 6 | 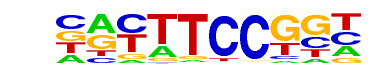 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-53 | -1.227e+02 | 0.0000 | 489.0 | 21.81% | 79.9 | 10.65% | motif file (matrix) |
pdf |
| 7 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-49 | -1.140e+02 | 0.0000 | 354.0 | 15.79% | 50.3 | 6.70% | motif file (matrix) |
pdf |
| 8 |  | ETS(ETS)/Promoter/Homer | 1e-47 | -1.094e+02 | 0.0000 | 183.0 | 8.16% | 17.4 | 2.31% | motif file (matrix) |
pdf |
| 9 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-38 | -8.779e+01 | 0.0000 | 60.0 | 2.68% | 2.3 | 0.30% | motif file (matrix) |
pdf |
| 10 | 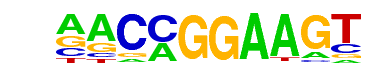 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-36 | -8.370e+01 | 0.0000 | 487.0 | 21.72% | 91.7 | 12.22% | motif file (matrix) |
pdf |
| 11 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-35 | -8.124e+01 | 0.0000 | 360.0 | 16.06% | 60.1 | 8.02% | motif file (matrix) |
pdf |
| 12 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-28 | -6.632e+01 | 0.0000 | 103.0 | 4.59% | 9.5 | 1.27% | motif file (matrix) |
pdf |
| 13 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-26 | -6.165e+01 | 0.0000 | 144.0 | 6.42% | 17.0 | 2.27% | motif file (matrix) |
pdf |
| 14 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.731e+01 | 0.0000 | 279.0 | 12.44% | 48.9 | 6.52% | motif file (matrix) |
pdf |
| 15 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-24 | -5.689e+01 | 0.0000 | 35.0 | 1.56% | 1.6 | 0.22% | motif file (matrix) |
pdf |
| 16 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-24 | -5.684e+01 | 0.0000 | 46.0 | 2.05% | 2.4 | 0.31% | motif file (matrix) |
pdf |
| 17 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-24 | -5.625e+01 | 0.0000 | 365.0 | 16.28% | 70.3 | 9.38% | motif file (matrix) |
pdf |
| 18 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-23 | -5.493e+01 | 0.0000 | 221.0 | 9.86% | 35.7 | 4.76% | motif file (matrix) |
pdf |
| 19 |  | E2F(E2F)/Cell-Cycle-Exp/Homer | 1e-22 | -5.197e+01 | 0.0000 | 33.0 | 1.47% | 0.3 | 0.04% | motif file (matrix) |
pdf |
| 20 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-22 | -5.197e+01 | 0.0000 | 33.0 | 1.47% | 0.3 | 0.04% | motif file (matrix) |
pdf |
| 21 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-22 | -5.116e+01 | 0.0000 | 417.0 | 18.60% | 86.9 | 11.59% | motif file (matrix) |
pdf |
| 22 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-20 | -4.761e+01 | 0.0000 | 245.0 | 10.93% | 43.4 | 5.79% | motif file (matrix) |
pdf |
| 23 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-19 | -4.391e+01 | 0.0000 | 80.0 | 3.57% | 8.7 | 1.16% | motif file (matrix) |
pdf |
| 24 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-17 | -4.101e+01 | 0.0000 | 38.0 | 1.69% | 2.8 | 0.38% | motif file (matrix) |
pdf |
| 25 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-17 | -3.921e+01 | 0.0000 | 373.0 | 16.64% | 80.3 | 10.70% | motif file (matrix) |
pdf |
| 26 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-16 | -3.705e+01 | 0.0000 | 85.0 | 3.79% | 10.0 | 1.33% | motif file (matrix) |
pdf |
| 27 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-15 | -3.648e+01 | 0.0000 | 320.0 | 14.27% | 67.9 | 9.05% | motif file (matrix) |
pdf |
| 28 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-15 | -3.622e+01 | 0.0000 | 154.0 | 6.87% | 25.9 | 3.45% | motif file (matrix) |
pdf |
| 29 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-15 | -3.592e+01 | 0.0000 | 352.0 | 15.70% | 76.7 | 10.23% | motif file (matrix) |
pdf |
| 30 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-14 | -3.452e+01 | 0.0000 | 562.0 | 25.07% | 137.2 | 18.29% | motif file (matrix) |
pdf |
| 31 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-14 | -3.366e+01 | 0.0000 | 526.0 | 23.46% | 127.6 | 17.01% | motif file (matrix) |
pdf |
| 32 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-14 | -3.334e+01 | 0.0000 | 532.0 | 23.73% | 129.7 | 17.29% | motif file (matrix) |
pdf |
| 33 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-13 | -3.189e+01 | 0.0000 | 53.0 | 2.36% | 5.9 | 0.79% | motif file (matrix) |
pdf |
| 34 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-13 | -3.150e+01 | 0.0000 | 314.0 | 14.01% | 68.5 | 9.14% | motif file (matrix) |
pdf |
| 35 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-13 | -3.123e+01 | 0.0000 | 503.0 | 22.44% | 122.9 | 16.38% | motif file (matrix) |
pdf |
| 36 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-12 | -2.960e+01 | 0.0000 | 115.0 | 5.13% | 18.3 | 2.44% | motif file (matrix) |
pdf |
| 37 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-12 | -2.867e+01 | 0.0000 | 290.0 | 12.93% | 63.1 | 8.41% | motif file (matrix) |
pdf |
| 38 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-12 | -2.857e+01 | 0.0000 | 421.0 | 18.78% | 100.6 | 13.42% | motif file (matrix) |
pdf |
| 39 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-12 | -2.856e+01 | 0.0000 | 164.0 | 7.31% | 30.3 | 4.05% | motif file (matrix) |
pdf |
| 40 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-12 | -2.847e+01 | 0.0000 | 91.0 | 4.06% | 13.9 | 1.86% | motif file (matrix) |
pdf |
| 41 |  | PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-12 | -2.831e+01 | 0.0000 | 100.0 | 4.46% | 16.0 | 2.13% | motif file (matrix) |
pdf |
| 42 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-12 | -2.795e+01 | 0.0000 | 179.0 | 7.98% | 34.9 | 4.66% | motif file (matrix) |
pdf |
| 43 |  | ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER | 1e-11 | -2.687e+01 | 0.0000 | 30.0 | 1.34% | 2.6 | 0.34% | motif file (matrix) |
pdf |
| 44 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-11 | -2.567e+01 | 0.0000 | 400.0 | 17.84% | 96.7 | 12.90% | motif file (matrix) |
pdf |
| 45 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-11 | -2.550e+01 | 0.0000 | 139.0 | 6.20% | 25.2 | 3.36% | motif file (matrix) |
pdf |
| 46 |  | NFY(CCAAT)/Promoter/Homer | 1e-10 | -2.436e+01 | 0.0000 | 196.0 | 8.74% | 40.5 | 5.40% | motif file (matrix) |
pdf |
| 47 |  | RBPJ:Ebox/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-10 | -2.418e+01 | 0.0000 | 86.0 | 3.84% | 13.9 | 1.86% | motif file (matrix) |
pdf |
| 48 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-9 | -2.277e+01 | 0.0000 | 193.0 | 8.61% | 40.5 | 5.41% | motif file (matrix) |
pdf |
| 49 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-9 | -2.231e+01 | 0.0000 | 294.0 | 13.11% | 68.3 | 9.11% | motif file (matrix) |
pdf |
| 50 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-9 | -2.169e+01 | 0.0000 | 172.0 | 7.67% | 35.9 | 4.79% | motif file (matrix) |
pdf |
| 51 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-9 | -2.150e+01 | 0.0000 | 164.0 | 7.31% | 33.6 | 4.48% | motif file (matrix) |
pdf |
| 52 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-8 | -2.047e+01 | 0.0000 | 207.0 | 9.23% | 45.8 | 6.11% | motif file (matrix) |
pdf |
| 53 | 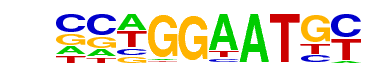 | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-8 | -2.037e+01 | 0.0000 | 261.0 | 11.64% | 60.7 | 8.10% | motif file (matrix) |
pdf |
| 54 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-8 | -1.984e+01 | 0.0000 | 202.0 | 9.01% | 44.7 | 5.97% | motif file (matrix) |
pdf |
| 55 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-8 | -1.949e+01 | 0.0000 | 487.0 | 21.72% | 127.7 | 17.02% | motif file (matrix) |
pdf |
| 56 |  | TATA-Box(TBP)/Promoter/Homer | 1e-8 | -1.902e+01 | 0.0000 | 409.0 | 18.24% | 104.1 | 13.88% | motif file (matrix) |
pdf |
| 57 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-8 | -1.868e+01 | 0.0000 | 264.0 | 11.78% | 62.8 | 8.38% | motif file (matrix) |
pdf |
| 58 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.771e+01 | 0.0000 | 134.0 | 5.98% | 27.2 | 3.62% | motif file (matrix) |
pdf |
| 59 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-7 | -1.762e+01 | 0.0000 | 24.0 | 1.07% | 2.3 | 0.30% | motif file (matrix) |
pdf |
| 60 | 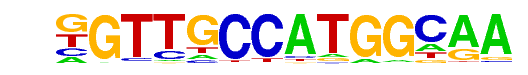 | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-7 | -1.643e+01 | 0.0000 | 63.0 | 2.81% | 10.1 | 1.35% | motif file (matrix) |
pdf |
| 61 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-6 | -1.586e+01 | 0.0000 | 671.0 | 29.93% | 188.9 | 25.19% | motif file (matrix) |
pdf |
| 62 | 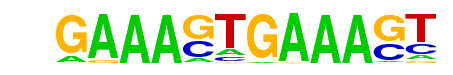 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-6 | -1.519e+01 | 0.0000 | 57.0 | 2.54% | 9.2 | 1.23% | motif file (matrix) |
pdf |
| 63 |  | E-box(HLH)/Promoter/Homer | 1e-6 | -1.512e+01 | 0.0000 | 28.0 | 1.25% | 3.5 | 0.46% | motif file (matrix) |
pdf |
| 64 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-6 | -1.493e+01 | 0.0000 | 43.0 | 1.92% | 7.0 | 0.93% | motif file (matrix) |
pdf |
| 65 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.483e+01 | 0.0000 | 219.0 | 9.77% | 52.4 | 6.99% | motif file (matrix) |
pdf |
| 66 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-6 | -1.480e+01 | 0.0000 | 458.0 | 20.43% | 123.7 | 16.50% | motif file (matrix) |
pdf |
| 67 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-6 | -1.478e+01 | 0.0000 | 435.0 | 19.40% | 116.9 | 15.58% | motif file (matrix) |
pdf |
| 68 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-6 | -1.477e+01 | 0.0000 | 33.0 | 1.47% | 4.7 | 0.62% | motif file (matrix) |
pdf |
| 69 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-5 | -1.352e+01 | 0.0000 | 46.0 | 2.05% | 7.8 | 1.04% | motif file (matrix) |
pdf |
| 70 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-5 | -1.290e+01 | 0.0000 | 102.0 | 4.55% | 21.9 | 2.92% | motif file (matrix) |
pdf |
| 71 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-5 | -1.267e+01 | 0.0000 | 227.0 | 10.12% | 56.5 | 7.53% | motif file (matrix) |
pdf |
| 72 | 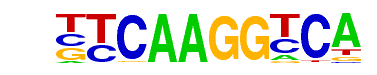 | Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer | 1e-5 | -1.265e+01 | 0.0000 | 124.0 | 5.53% | 27.5 | 3.67% | motif file (matrix) |
pdf |
| 73 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.224e+01 | 0.0000 | 20.0 | 0.89% | 2.9 | 0.39% | motif file (matrix) |
pdf |
| 74 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-5 | -1.224e+01 | 0.0000 | 20.0 | 0.89% | 2.4 | 0.32% | motif file (matrix) |
pdf |
| 75 |  | Reverb(NR/DR2)/BLRP(RAW)-Reverba-ChIP-Seq(GSE45914)/Homer | 1e-5 | -1.224e+01 | 0.0000 | 20.0 | 0.89% | 2.4 | 0.32% | motif file (matrix) |
pdf |
| 76 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-5 | -1.191e+01 | 0.0000 | 211.0 | 9.41% | 52.6 | 7.02% | motif file (matrix) |
pdf |
| 77 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529/Homer | 1e-5 | -1.177e+01 | 0.0000 | 151.0 | 6.74% | 35.0 | 4.67% | motif file (matrix) |
pdf |
| 78 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-5 | -1.166e+01 | 0.0000 | 172.0 | 7.67% | 41.7 | 5.56% | motif file (matrix) |
pdf |
| 79 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -1.108e+01 | 0.0000 | 212.0 | 9.46% | 53.0 | 7.07% | motif file (matrix) |
pdf |
| 80 |  | GFX(?)/Promoter/Homer | 1e-4 | -1.108e+01 | 0.0000 | 13.0 | 0.58% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 81 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.101e+01 | 0.0000 | 19.0 | 0.85% | 2.7 | 0.36% | motif file (matrix) |
pdf |
| 82 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-4 | -1.101e+01 | 0.0000 | 19.0 | 0.85% | 2.6 | 0.35% | motif file (matrix) |
pdf |
| 83 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-4 | -1.101e+01 | 0.0000 | 19.0 | 0.85% | 2.6 | 0.35% | motif file (matrix) |
pdf |
| 84 |  | HOXD13(Homeobox)/Chicken-Hoxd13-ChIP-Seq(GSE38910)/Homer | 1e-4 | -1.035e+01 | 0.0001 | 379.0 | 16.90% | 104.4 | 13.92% | motif file (matrix) |
pdf |
| 85 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-4 | -1.006e+01 | 0.0001 | 58.0 | 2.59% | 11.2 | 1.50% | motif file (matrix) |
pdf |
| 86 |  | Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-4 | -9.927e+00 | 0.0001 | 81.0 | 3.61% | 17.6 | 2.35% | motif file (matrix) |
pdf |
| 87 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-4 | -9.868e+00 | 0.0001 | 377.0 | 16.82% | 104.0 | 13.87% | motif file (matrix) |
pdf |
| 88 |  | NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.500e+00 | 0.0002 | 95.0 | 4.24% | 21.0 | 2.81% | motif file (matrix) |
pdf |
| 89 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-4 | -9.313e+00 | 0.0002 | 76.0 | 3.39% | 16.7 | 2.23% | motif file (matrix) |
pdf |
| 90 |  | Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer | 1e-3 | -9.153e+00 | 0.0003 | 216.0 | 9.63% | 56.1 | 7.48% | motif file (matrix) |
pdf |
| 91 |  | PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-3 | -9.153e+00 | 0.0003 | 216.0 | 9.63% | 56.1 | 7.48% | motif file (matrix) |
pdf |
| 92 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-3 | -9.036e+00 | 0.0003 | 259.0 | 11.55% | 69.5 | 9.27% | motif file (matrix) |
pdf |
| 93 |  | CRE(bZIP)/Promoter/Homer | 1e-3 | -8.757e+00 | 0.0004 | 86.0 | 3.84% | 19.2 | 2.57% | motif file (matrix) |
pdf |
| 94 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.389e+00 | 0.0006 | 121.0 | 5.40% | 30.0 | 4.00% | motif file (matrix) |
pdf |
| 95 |  | STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer | 1e-2 | -6.878e+00 | 0.0027 | 161.0 | 7.18% | 43.0 | 5.73% | motif file (matrix) |
pdf |
| 96 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.643e+00 | 0.0033 | 116.0 | 5.17% | 30.0 | 4.00% | motif file (matrix) |
pdf |
| 97 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-2 | -6.627e+00 | 0.0034 | 246.0 | 10.97% | 68.4 | 9.12% | motif file (matrix) |
pdf |
| 98 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-2 | -6.593e+00 | 0.0034 | 102.0 | 4.55% | 25.9 | 3.45% | motif file (matrix) |
pdf |
| 99 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-2 | -6.581e+00 | 0.0034 | 24.0 | 1.07% | 4.1 | 0.54% | motif file (matrix) |
pdf |
| 100 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-2 | -6.469e+00 | 0.0038 | 77.0 | 3.43% | 18.3 | 2.44% | motif file (matrix) |
pdf |
| 101 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.164e+00 | 0.0051 | 185.0 | 8.25% | 50.4 | 6.72% | motif file (matrix) |
pdf |
| 102 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.890e+00 | 0.0067 | 537.0 | 23.95% | 161.7 | 21.57% | motif file (matrix) |
pdf |
| 103 |  | Hoxc9(Homeobox)/Ainv15-Hoxc9-ChIP-Seq(GSE21812)/Homer | 1e-2 | -5.700e+00 | 0.0080 | 140.0 | 6.24% | 37.5 | 5.00% | motif file (matrix) |
pdf |
| 104 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-2 | -5.557e+00 | 0.0091 | 64.0 | 2.85% | 15.1 | 2.01% | motif file (matrix) |
pdf |
| 105 |  | Srebp2(HLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.276e+00 | 0.0120 | 18.0 | 0.80% | 3.7 | 0.49% | motif file (matrix) |
pdf |
| 106 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -5.165e+00 | 0.0133 | 63.0 | 2.81% | 15.5 | 2.07% | motif file (matrix) |
pdf |
| 107 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -4.767e+00 | 0.0195 | 386.0 | 17.22% | 115.9 | 15.45% | motif file (matrix) |
pdf |
| 108 |  | GFY-Staf/Promoters/Homer | 1e-2 | -4.631e+00 | 0.0222 | 29.0 | 1.29% | 6.9 | 0.92% | motif file (matrix) |
pdf |