| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 | 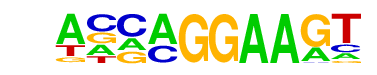 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-11 | -2.676e+01 | 0.0000 | 96.0 | 45.07% | 69.0 | 23.56% | motif file (matrix) |
pdf |
| 2 | 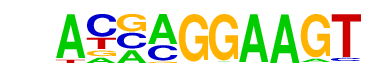 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-10 | -2.345e+01 | 0.0000 | 65.0 | 30.52% | 39.4 | 13.46% | motif file (matrix) |
pdf |
| 3 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-9 | -2.235e+01 | 0.0000 | 19.0 | 8.92% | 4.2 | 1.45% | motif file (matrix) |
pdf |
| 4 | 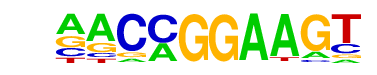 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-9 | -2.219e+01 | 0.0000 | 93.0 | 43.66% | 70.9 | 24.21% | motif file (matrix) |
pdf |
| 5 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-9 | -2.152e+01 | 0.0000 | 74.0 | 34.74% | 50.3 | 17.17% | motif file (matrix) |
pdf |
| 6 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-8 | -2.007e+01 | 0.0000 | 79.0 | 37.09% | 58.0 | 19.79% | motif file (matrix) |
pdf |
| 7 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.799e+01 | 0.0000 | 46.0 | 21.60% | 26.2 | 8.95% | motif file (matrix) |
pdf |
| 8 | 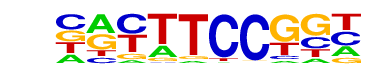 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-7 | -1.744e+01 | 0.0000 | 76.0 | 35.68% | 57.0 | 19.47% | motif file (matrix) |
pdf |
| 9 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-6 | -1.561e+01 | 0.0000 | 105.0 | 49.30% | 94.9 | 32.42% | motif file (matrix) |
pdf |
| 10 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.477e+01 | 0.0000 | 107.0 | 50.23% | 98.7 | 33.72% | motif file (matrix) |
pdf |
| 11 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-6 | -1.451e+01 | 0.0000 | 109.0 | 51.17% | 101.4 | 34.63% | motif file (matrix) |
pdf |
| 12 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-6 | -1.437e+01 | 0.0000 | 18.0 | 8.45% | 6.9 | 2.34% | motif file (matrix) |
pdf |
| 13 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.430e+01 | 0.0000 | 30.0 | 14.08% | 15.3 | 5.23% | motif file (matrix) |
pdf |
| 14 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-5 | -1.350e+01 | 0.0000 | 62.0 | 29.11% | 47.1 | 16.08% | motif file (matrix) |
pdf |
| 15 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.223e+01 | 0.0001 | 18.0 | 8.45% | 7.9 | 2.69% | motif file (matrix) |
pdf |
| 16 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.211e+01 | 0.0001 | 46.0 | 21.60% | 32.3 | 11.02% | motif file (matrix) |
pdf |
| 17 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.169e+01 | 0.0001 | 31.0 | 14.55% | 18.5 | 6.32% | motif file (matrix) |
pdf |
| 18 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -1.137e+01 | 0.0002 | 105.0 | 49.30% | 102.0 | 34.85% | motif file (matrix) |
pdf |
| 19 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-4 | -9.805e+00 | 0.0007 | 45.0 | 21.13% | 34.6 | 11.83% | motif file (matrix) |
pdf |
| 20 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-3 | -9.205e+00 | 0.0012 | 69.0 | 32.39% | 62.4 | 21.32% | motif file (matrix) |
pdf |
| 21 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -9.034e+00 | 0.0014 | 43.0 | 20.19% | 33.2 | 11.34% | motif file (matrix) |
pdf |
| 22 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.958e+00 | 0.0014 | 8.0 | 3.76% | 2.7 | 0.93% | motif file (matrix) |
pdf |
| 23 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-3 | -8.828e+00 | 0.0016 | 30.0 | 14.08% | 20.7 | 7.08% | motif file (matrix) |
pdf |
| 24 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-3 | -8.784e+00 | 0.0016 | 58.0 | 27.23% | 50.9 | 17.40% | motif file (matrix) |
pdf |
| 25 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-3 | -8.573e+00 | 0.0019 | 107.0 | 50.23% | 111.6 | 38.12% | motif file (matrix) |
pdf |
| 26 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.745e+00 | 0.0041 | 82.0 | 38.50% | 81.7 | 27.89% | motif file (matrix) |
pdf |
| 27 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -7.379e+00 | 0.0057 | 35.0 | 16.43% | 27.7 | 9.46% | motif file (matrix) |
pdf |
| 28 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.250e+00 | 0.0062 | 14.0 | 6.57% | 7.3 | 2.49% | motif file (matrix) |
pdf |
| 29 |  | NFY(CCAAT)/Promoter/Homer | 1e-3 | -7.240e+00 | 0.0062 | 44.0 | 20.66% | 37.8 | 12.93% | motif file (matrix) |
pdf |
| 30 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.215e+00 | 0.0062 | 27.0 | 12.68% | 19.4 | 6.64% | motif file (matrix) |
pdf |
| 31 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-3 | -7.206e+00 | 0.0062 | 7.0 | 3.29% | 2.5 | 0.86% | motif file (matrix) |
pdf |
| 32 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-3 | -7.113e+00 | 0.0063 | 91.0 | 42.72% | 94.8 | 32.38% | motif file (matrix) |
pdf |
| 33 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-3 | -7.083e+00 | 0.0063 | 132.0 | 61.97% | 149.4 | 51.02% | motif file (matrix) |
pdf |
| 34 |  | ETS(ETS)/Promoter/Homer | 1e-2 | -6.646e+00 | 0.0094 | 18.0 | 8.45% | 11.6 | 3.97% | motif file (matrix) |
pdf |
| 35 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.548e+00 | 0.0101 | 49.0 | 23.00% | 44.3 | 15.12% | motif file (matrix) |
pdf |
| 36 | 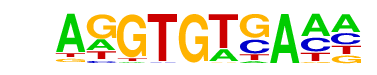 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-2 | -6.533e+00 | 0.0101 | 65.0 | 30.52% | 64.0 | 21.85% | motif file (matrix) |
pdf |
| 37 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.483e+00 | 0.0102 | 21.0 | 9.86% | 14.1 | 4.81% | motif file (matrix) |
pdf |
| 38 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-2 | -6.445e+00 | 0.0103 | 23.0 | 10.80% | 16.5 | 5.64% | motif file (matrix) |
pdf |
| 39 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.383e+00 | 0.0107 | 12.0 | 5.63% | 6.5 | 2.23% | motif file (matrix) |
pdf |
| 40 |  | STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer | 1e-2 | -6.328e+00 | 0.0110 | 46.0 | 21.60% | 41.0 | 14.01% | motif file (matrix) |
pdf |
| 41 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-2 | -6.010e+00 | 0.0147 | 80.0 | 37.56% | 83.2 | 28.42% | motif file (matrix) |
pdf |
| 42 |  | c-Myc/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.825e+00 | 0.0173 | 28.0 | 13.15% | 22.8 | 7.78% | motif file (matrix) |
pdf |
| 43 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-2 | -5.677e+00 | 0.0196 | 22.0 | 10.33% | 16.2 | 5.55% | motif file (matrix) |
pdf |
| 44 |  | NFkB-p50,p52(RHD)/p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-2 | -5.586e+00 | 0.0210 | 6.0 | 2.82% | 2.8 | 0.94% | motif file (matrix) |
pdf |
| 45 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-2 | -5.372e+00 | 0.0254 | 31.0 | 14.55% | 26.9 | 9.20% | motif file (matrix) |
pdf |
| 46 |  | Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-2 | -5.304e+00 | 0.0266 | 11.0 | 5.16% | 7.0 | 2.38% | motif file (matrix) |
pdf |
| 47 |  | CHR/Cell-Cycle-Exp/Homer | 1e-2 | -5.286e+00 | 0.0266 | 50.0 | 23.47% | 49.0 | 16.72% | motif file (matrix) |
pdf |
| 48 |  | Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer | 1e-2 | -5.286e+00 | 0.0266 | 50.0 | 23.47% | 48.7 | 16.63% | motif file (matrix) |
pdf |
| 49 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-2 | -5.234e+00 | 0.0268 | 102.0 | 47.89% | 114.1 | 38.95% | motif file (matrix) |
pdf |
| 50 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -5.159e+00 | 0.0283 | 155.0 | 72.77% | 188.3 | 64.31% | motif file (matrix) |
pdf |
| 51 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-2 | -5.104e+00 | 0.0293 | 66.0 | 30.99% | 68.7 | 23.45% | motif file (matrix) |
pdf |
| 52 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-2 | -5.062e+00 | 0.0300 | 13.0 | 6.10% | 8.7 | 2.98% | motif file (matrix) |
pdf |
| 53 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.981e+00 | 0.0319 | 22.0 | 10.33% | 17.2 | 5.88% | motif file (matrix) |
pdf |
| 54 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-2 | -4.972e+00 | 0.0319 | 7.0 | 3.29% | 3.3 | 1.12% | motif file (matrix) |
pdf |
| 55 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-2 | -4.972e+00 | 0.0319 | 7.0 | 3.29% | 3.3 | 1.12% | motif file (matrix) |
pdf |
| 56 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-2 | -4.972e+00 | 0.0319 | 7.0 | 3.29% | 3.1 | 1.05% | motif file (matrix) |
pdf |
| 57 |  | RBPJ:Ebox/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -4.910e+00 | 0.0319 | 18.0 | 8.45% | 13.5 | 4.60% | motif file (matrix) |
pdf |
| 58 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-2 | -4.903e+00 | 0.0319 | 45.0 | 21.13% | 43.2 | 14.75% | motif file (matrix) |
pdf |
| 59 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-2 | -4.829e+00 | 0.0333 | 44.0 | 20.66% | 42.8 | 14.63% | motif file (matrix) |
pdf |
| 60 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.617e+00 | 0.0405 | 35.0 | 16.43% | 32.7 | 11.17% | motif file (matrix) |
pdf |























































































































