| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 | 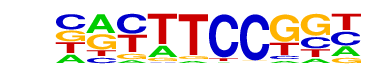 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-12 | -2.949e+01 | 0.0000 | 1268.0 | 34.24% | 1130.9 | 28.74% | motif file (matrix) |
pdf |
| 2 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-11 | -2.654e+01 | 0.0000 | 678.0 | 18.31% | 559.5 | 14.22% | motif file (matrix) |
pdf |
| 3 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-11 | -2.636e+01 | 0.0000 | 1300.0 | 35.11% | 1176.0 | 29.88% | motif file (matrix) |
pdf |
| 4 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-10 | -2.434e+01 | 0.0000 | 1988.0 | 53.69% | 1900.1 | 48.29% | motif file (matrix) |
pdf |
| 5 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-9 | -2.175e+01 | 0.0000 | 2037.0 | 55.01% | 1965.3 | 49.94% | motif file (matrix) |
pdf |
| 6 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-8 | -2.014e+01 | 0.0000 | 1048.0 | 28.30% | 947.8 | 24.09% | motif file (matrix) |
pdf |
| 7 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-7 | -1.798e+01 | 0.0000 | 2002.0 | 54.06% | 1948.4 | 49.51% | motif file (matrix) |
pdf |
| 8 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-7 | -1.717e+01 | 0.0000 | 510.0 | 13.77% | 429.9 | 10.93% | motif file (matrix) |
pdf |
| 9 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.667e+01 | 0.0000 | 1091.0 | 29.46% | 1007.3 | 25.60% | motif file (matrix) |
pdf |
| 10 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.572e+01 | 0.0000 | 763.0 | 20.60% | 682.3 | 17.34% | motif file (matrix) |
pdf |
| 11 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-6 | -1.564e+01 | 0.0000 | 1672.0 | 45.15% | 1613.5 | 41.00% | motif file (matrix) |
pdf |
| 12 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.321e+01 | 0.0000 | 273.0 | 7.37% | 218.6 | 5.56% | motif file (matrix) |
pdf |
| 13 | 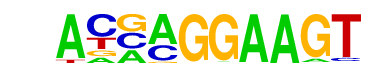 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-5 | -1.247e+01 | 0.0001 | 924.0 | 24.95% | 860.9 | 21.88% | motif file (matrix) |
pdf |
| 14 | 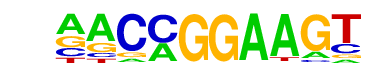 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-5 | -1.233e+01 | 0.0001 | 1406.0 | 37.97% | 1356.4 | 34.47% | motif file (matrix) |
pdf |
| 15 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-5 | -1.216e+01 | 0.0001 | 1585.0 | 42.80% | 1544.7 | 39.25% | motif file (matrix) |
pdf |
| 16 | 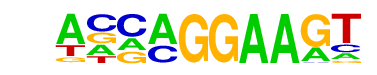 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-5 | -1.181e+01 | 0.0001 | 1466.0 | 39.59% | 1422.1 | 36.14% | motif file (matrix) |
pdf |
| 17 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-4 | -1.150e+01 | 0.0001 | 1126.0 | 30.41% | 1072.1 | 27.24% | motif file (matrix) |
pdf |
| 18 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.095e+01 | 0.0002 | 1028.0 | 27.76% | 975.9 | 24.80% | motif file (matrix) |
pdf |
| 19 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-4 | -9.981e+00 | 0.0006 | 382.0 | 10.32% | 333.4 | 8.47% | motif file (matrix) |
pdf |
| 20 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-4 | -9.931e+00 | 0.0006 | 914.0 | 24.68% | 865.4 | 21.99% | motif file (matrix) |
pdf |
| 21 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-4 | -9.587e+00 | 0.0008 | 562.0 | 15.18% | 512.1 | 13.01% | motif file (matrix) |
pdf |
| 22 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-3 | -8.828e+00 | 0.0016 | 1019.0 | 27.52% | 980.4 | 24.92% | motif file (matrix) |
pdf |
| 23 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-3 | -8.638e+00 | 0.0019 | 106.0 | 2.86% | 78.2 | 1.99% | motif file (matrix) |
pdf |
| 24 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-3 | -8.416e+00 | 0.0023 | 1172.0 | 31.65% | 1141.5 | 29.01% | motif file (matrix) |
pdf |
| 25 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-3 | -8.399e+00 | 0.0023 | 452.0 | 12.21% | 409.3 | 10.40% | motif file (matrix) |
pdf |
| 26 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.898e+00 | 0.0035 | 727.0 | 19.63% | 688.8 | 17.50% | motif file (matrix) |
pdf |
| 27 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-3 | -7.719e+00 | 0.0040 | 303.0 | 8.18% | 266.7 | 6.78% | motif file (matrix) |
pdf |
| 28 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.491e+00 | 0.0049 | 986.0 | 26.63% | 957.0 | 24.32% | motif file (matrix) |
pdf |
| 29 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.838e+00 | 0.0091 | 566.0 | 15.28% | 532.6 | 13.53% | motif file (matrix) |
pdf |
| 30 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-2 | -6.836e+00 | 0.0091 | 369.0 | 9.96% | 335.0 | 8.51% | motif file (matrix) |
pdf |
| 31 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-2 | -6.756e+00 | 0.0092 | 101.0 | 2.73% | 78.6 | 2.00% | motif file (matrix) |
pdf |
| 32 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -6.699e+00 | 0.0095 | 2699.0 | 72.89% | 2779.2 | 70.63% | motif file (matrix) |
pdf |
| 33 |  | NFY(CCAAT)/Promoter/Homer | 1e-2 | -6.582e+00 | 0.0103 | 715.0 | 19.31% | 685.2 | 17.41% | motif file (matrix) |
pdf |
| 34 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-2 | -6.015e+00 | 0.0177 | 1742.0 | 47.04% | 1760.7 | 44.74% | motif file (matrix) |
pdf |
| 35 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.992e+00 | 0.0177 | 504.0 | 13.61% | 475.1 | 12.07% | motif file (matrix) |
pdf |
| 36 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.945e+00 | 0.0179 | 563.0 | 15.20% | 535.4 | 13.61% | motif file (matrix) |
pdf |
| 37 |  | CHR/Cell-Cycle-Exp/Homer | 1e-2 | -5.945e+00 | 0.0179 | 762.0 | 20.58% | 738.0 | 18.75% | motif file (matrix) |
pdf |
| 38 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.873e+00 | 0.0182 | 204.0 | 5.51% | 178.6 | 4.54% | motif file (matrix) |
pdf |
| 39 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.701e+00 | 0.0211 | 1482.0 | 40.02% | 1489.2 | 37.85% | motif file (matrix) |
pdf |
| 40 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-2 | -5.639e+00 | 0.0219 | 703.0 | 18.98% | 680.9 | 17.30% | motif file (matrix) |
pdf |
| 41 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-2 | -5.611e+00 | 0.0219 | 649.0 | 17.53% | 625.4 | 15.89% | motif file (matrix) |
pdf |
| 42 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-2 | -5.589e+00 | 0.0219 | 1498.0 | 40.45% | 1507.4 | 38.31% | motif file (matrix) |
pdf |
| 43 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-2 | -5.511e+00 | 0.0231 | 190.0 | 5.13% | 166.9 | 4.24% | motif file (matrix) |
pdf |
| 44 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.393e+00 | 0.0254 | 2284.0 | 61.68% | 2345.0 | 59.59% | motif file (matrix) |
pdf |
| 45 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-2 | -5.391e+00 | 0.0254 | 130.0 | 3.51% | 109.8 | 2.79% | motif file (matrix) |
pdf |
| 46 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.219e+00 | 0.0289 | 475.0 | 12.83% | 451.5 | 11.47% | motif file (matrix) |
pdf |
| 47 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.131e+00 | 0.0309 | 411.0 | 11.10% | 387.8 | 9.86% | motif file (matrix) |
pdf |
| 48 |  | HOXD13(Homeobox)/Chicken-Hoxd13-ChIP-Seq(GSE38910)/Homer | 1e-2 | -5.130e+00 | 0.0309 | 1236.0 | 33.38% | 1237.2 | 31.44% | motif file (matrix) |
pdf |
| 49 | 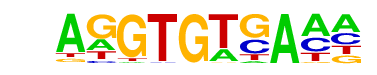 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-2 | -5.116e+00 | 0.0309 | 1047.0 | 28.27% | 1040.9 | 26.45% | motif file (matrix) |
pdf |
| 50 |  | Srebp1a(HLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.030e+00 | 0.0322 | 245.0 | 6.62% | 222.0 | 5.64% | motif file (matrix) |
pdf |
| 51 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.987e+00 | 0.0329 | 140.0 | 3.78% | 120.3 | 3.06% | motif file (matrix) |
pdf |
| 52 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-2 | -4.813e+00 | 0.0384 | 738.0 | 19.93% | 723.0 | 18.37% | motif file (matrix) |
pdf |
| 53 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-2 | -4.811e+00 | 0.0384 | 1634.0 | 44.13% | 1659.7 | 42.18% | motif file (matrix) |
pdf |
| 54 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -4.707e+00 | 0.0411 | 119.0 | 3.21% | 101.3 | 2.57% | motif file (matrix) |
pdf |
| 55 |  | Reverb(NR/DR2)/BLRP(RAW)-Reverba-ChIP-Seq(GSE45914)/Homer | 1e-2 | -4.634e+00 | 0.0434 | 151.0 | 4.08% | 132.2 | 3.36% | motif file (matrix) |
pdf |













































































































